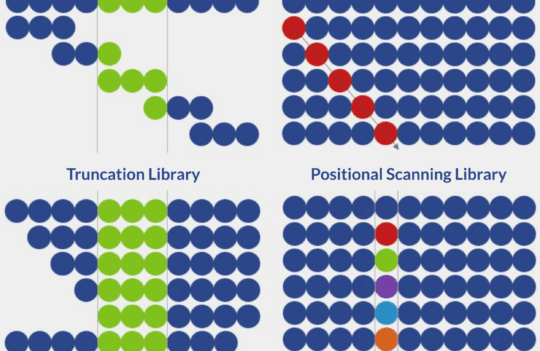
Peptide synthesis
Peptide synthesis
How synthetic peptide libraries are being used to boost high-throughput protein interaction analysis
Much has been published about screening peptide libraries via biological display techniques such as phage display. But what do we know about screening synthetic peptide libraries in vitro? Conventional methods for scanning these libraries, although optimal for high-throughput interaction analysis, are often limited in terms of diversity and sensitivity. But in the race to overcome this limitation, numerous new methodologies are popularizing the use of increasingly diverse synthetic libraries for multiple types of studies.
- The value of peptides for protein interaction analysis
- High-throughput screening of peptide libraries
- ELISA for protein interaction analysis
- Cytotoxic and antimicrobial detection assays for protein interaction studies
- Affinity-based selection coupled with mass spectrometry for protein interaction analysis
- Concluding remarks
- References
The value of peptides for protein interaction analysis
Peptides represent specific domains or fragments of larger proteins. Their major strength is their ability to bind large molecules and disrupt or facilitate biological processes, a crucial property for cell signalling and drug development applications. Besides their flexibility, peptides are considerably smaller than proteins and thus easier, quicker, and more cost-effective to produce.
As a result of their size, they can also cross membranes more efficiently and reach targets that would otherwise remain undruggable. Additionally, they are compatible with many tools used for high-throughput protein interaction analysis.
For this type of analysis, the rapid synthesis of vast collections of peptides is a must. These collections, also known as peptide libraries, can contain multiple random or targeted substitutions and modifications, allowing researchers to understand which residues of a protein are essential for interaction and which residues could be replaced to enhance stability and bioactivity.
High-throughput screening of peptide libraries
Protein interaction analysis comprises studies of protein-protein affinity. In these studies, peptides may be immobilized on different solid supports or tagged with fluorescent or other types of dyes and screened in solution.
The major difference between the two kinds of approaches lies in the final structure of the peptide. When peptides are immobilized on solid supports, their structure and bioactivity may be modified. As a result, these experiments often produce data that fails to reflect the real bioactivity of a peptide in solution.
Thus, the use of free peptides for high-throughput protein interaction analysis is preferred. However, these assays are not without their challenges. Indeed, in solution, it is often harder to miniaturize and automate assays while ensuring they remain highly sensitive and robust.
With these requirements in mind, many assays have been developed to allow high-throughput protein interaction analysis.
ELISA for protein interaction analysis
ELISA remains the most popular format for protein-protein interaction studies. Conventional ELISA uses 96-well plates, but newer assays have miniaturized it further allowing the use of 384-well plates in fully automated setups.
The most common type of detection is colorimetric, but ELISA can be easily adapted for fluorescent detection. In both cases, as the volume of the assay decreases, the need for very sensitive detection methods increases.
Cytotoxic and antimicrobial detection assays for protein interaction studies
Cell-based cytotoxic assays or antimicrobial assays remain useful when studying libraries of peptides with potential cytotoxic or antimicrobial activity. In a conventional sense, this type of assay is harder to miniaturize and automate.
However, newer technologies are constantly being developed for the rapid prospection of vast collections of peptide sequences. The new approaches rely on the use of 96 or 384-well microtiter plates and measure cell viability or growth over a set period of time.
Affinity-based selection coupled with mass spectrometry for protein interaction analysis
Affinity selection coupled with mass spectrometry (MS) is one of the most recent and promising alternatives to the conventional methods used in protein interaction studies. This method is proving to be especially useful to screen fully randomized sequences of peptides, the most diverse type of synthetic peptide libraries.
This type of method is based on the interaction between free peptides (binders) and one or several ligands immobilized on a solid surface. Ligands can be immobilized on resins (classical affinity chromatography) or magnetic beads. In both cases, these compounds are used to capture and enrich synthetic peptide libraries and subsequently sequence the most bioactive compounds via liquid chromatography coupled with MS.
What makes these methods so powerful? MS-based methods of protein interaction analysis have finally achieved the level of sensitivity previously restricted to biological assays (i.e., phage, bacterial, or ribosomal display). In other words, screening synthetic peptide libraries as diverse as genetically-encoded libraries (up to 109) is finally within reach.
Not only are these methods as sensitive as biological assays, but they also present an important advantage over genetically-encoded libraries. Unlike biological libraries, synthetic libraries are not restricted to natural and unmodified amino acids. For this reason, MS-based screening of modified peptides has the potential to expedite peptide drug and biomarker discovery efforts.
Concluding remarks
Synthetic peptide libraries allow the study of vast collections of randomized and modified peptides. Up until recently, the use of these libraries to study protein interactions had been limited to low-throughput screening methods. But with the rise of increasingly sensitive and miniaturized technologies, the use of synthetic libraries has been gaining ground over biological display for these applications.
This change is currently propelled by the further miniaturization of conventional ELISA and cell-based activity assays where the use of 384-well plates and automatized step-ups has been standardized. Moreover, MS-based methodologies coupled with a peptide enrichment step have finally reached the level of diversity previously restricted to biological display.
References
- Blikstad, C. and Ivarsson, Y. High-throughput methods for identification of protein-protein interactions involving short linear motifs. Cell Commun Signal. 2015; 13: 38. doi: 10.1186/s12964-015-0116-8
- Falciani, C. et al. Bioactive peptides from libraries. Chem Biol. 2005; 12(4): 417-426. doi: 10.1016/j.chembiol.2005.02.009
- Quartararo, A. J., et al. Ultra-large chemical libraries for the discovery of high-affinity peptide binders. Nat Commun. 2020; 11: 3183. doi: 10.1038/s41467-020-16920-3