 Peptide synthesis
Peptide synthesis
All you need to know about peptide library design
The design of peptide libraries entails the creation of collections of peptide sequences to unravel the link between sequence and bioactivity. Peptide libraries can accommodate a vast diversity of peptides or be restricted to only a few sequences, depending on the design strategy. On a fundamental level, peptide libraries are also classified as synthetic or genetically-encoded, depending on the method used for their production. In this article, we will analyze the suitability of each type of library for biomarker discovery, drug development, and enzyme and antibody optimization.
- What is the purpose of designing peptide libraries?
- Genetically-encoded versus synthetic peptide libraries
- How are peptide libraries designed?
- Overlapping peptide libraries
- Alanine scanning peptide libraries
- Truncation peptide libraries
- Positional scanning peptide libraries
- Random peptide libraries
- Scrambled peptide libraries
- References
What is the purpose of designing peptide libraries?
Peptides are the building blocks and catalysts of important biological processes. They occur naturally, serving as ligands or acting as disruptors or modifiers of biological processes. Due to these properties, peptides have become invaluable for biopharmaceutical, cosmetic, and food industries, and continue to find new and exciting applications.
But native peptides are intrinsically limited. Without optimization, these molecules tend to aggregate under physiological conditions, limiting their reactivity and usefulness. Additionally, not all peptide domains are equally important for biological activity, making sequence optimization and extensive characterization key to their success in different applications.
To attain the best stability and activity, researchers have leveraged the simplicity of solid-phase peptide synthesis and devised simple strategies to identify key and bioactive regions and residues within a protein. These strategies comprise the design of combinatorial peptide libraries that are easily produced and screened using high-throughput technologies.
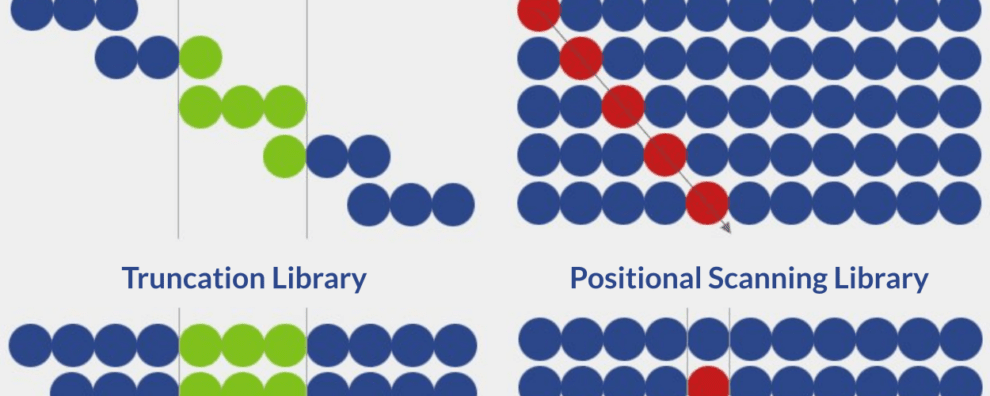
There are currently six distinct strategies for the design of peptide libraries:
- Overlapping library
- Alanine scanning library
- Truncation library
- Positional scanning library
- Random library
- Scrambled library
These strategies for peptide library design allow the discovery of new targets for treatment, biomarkers for diagnostics, or markers for environmental and food monitoring applications. Choosing the right library design dictates how informative a library screening experiment will be. But before analyzing the advantages and limitations of each type of design, we will dive into the different methods of peptide library synthesis.
Genetically-encoded versus synthetic peptide libraries
Combinatorial peptide libraries are collections of peptides with a defined length and systematic combinations of amino acids. Depending on the method used for their production, libraries can be defined as synthetic or genetically-encoded. The latter is often referred to as phage display peptide libraries.
As the name indicates, synthetic libraries are produced using amino acids as building blocks to create new polypeptide chains in a stepwise manner. The most widespread method of peptide synthesis is the solid-phase process, mediated by the Fmoc (9-fluorenylmethoxycarbonyl) N-terminus protective group. Once synthesized, these libraries can be screened in solution or immobilized on a solid support using methods such as ELISA or peptide microarrays.
In contrast, phage-displayed peptide libraries are commonly produced by synthesizing the nucleic acid sequence corresponding to each peptide, cloning it into a phagemid, and displaying it on the surface of Escherichia coli’s filamentous phage M13. Unlike synthetic libraries, these libraries are built for quick screening in solution. Promising ligands are easily captured by binding them to antibodies, non-antibody proteins (e.g., enzymes), small molecules, or even whole cells.
The advantages and limitations of each type of peptide library production process are listed below:
| Advantages | Limitations | |
|---|---|---|
| Genetically-encoded or phage display peptide libraries | Not limited by length
Accommodate high diversity of peptides (108-1013) Ease of library amplification and replication in bacteria Biopanning of peptides can be done against a number of different molecules or cells Phage particles are amenable to variations in culture conditions |
Limited to natural amino acids
Biological pressures can create bias and skew library diversity Screening of peptides is not quantitative Promising lead peptides need to be sequenced The process of screening and selecting promising binders is labor-intensive |
| Synthetic peptide libraries | Greater flexibility in terms of peptide library design
Compatible with unnatural, modified, cyclic, and D-amino acids Promising peptide sequences can be rapidly optimized (without needing to be sequenced) Shorter lead times in comparison to phage display library production and screening Greater potential to identify stable peptide sequences Free from biological bias |
Less diverse than genetically-encoded libraries (typically 106) |
Synthetic libraries are compatible with a broader diversity of chemical structures. They are fast to produce and screen. However, they are often limited to a lower diversity of sequences in comparison to genetically-encoded libraries.
But thanks to the development of high-resolution and high-throughput screening techniques, this is quickly changing. Chemical libraries are increasingly screened using techniques such as affinity selection-mass spectrometry (AS-MS), which represent one of the most promising alternatives to conventional ELISA or microarray assays.
How are peptide libraries designed?
Most peptide libraries, either synthetic or genetically-encoded, are designed by identifying bioactive regions in native proteins. Currently, some online tools can be used to predict these regions, including the Bioware web server hosted by the University College Dublin.
After selecting the regions predicted to be bioactive, researchers must select an appropriate peptide library design according to the purpose of the experiment and the final application.
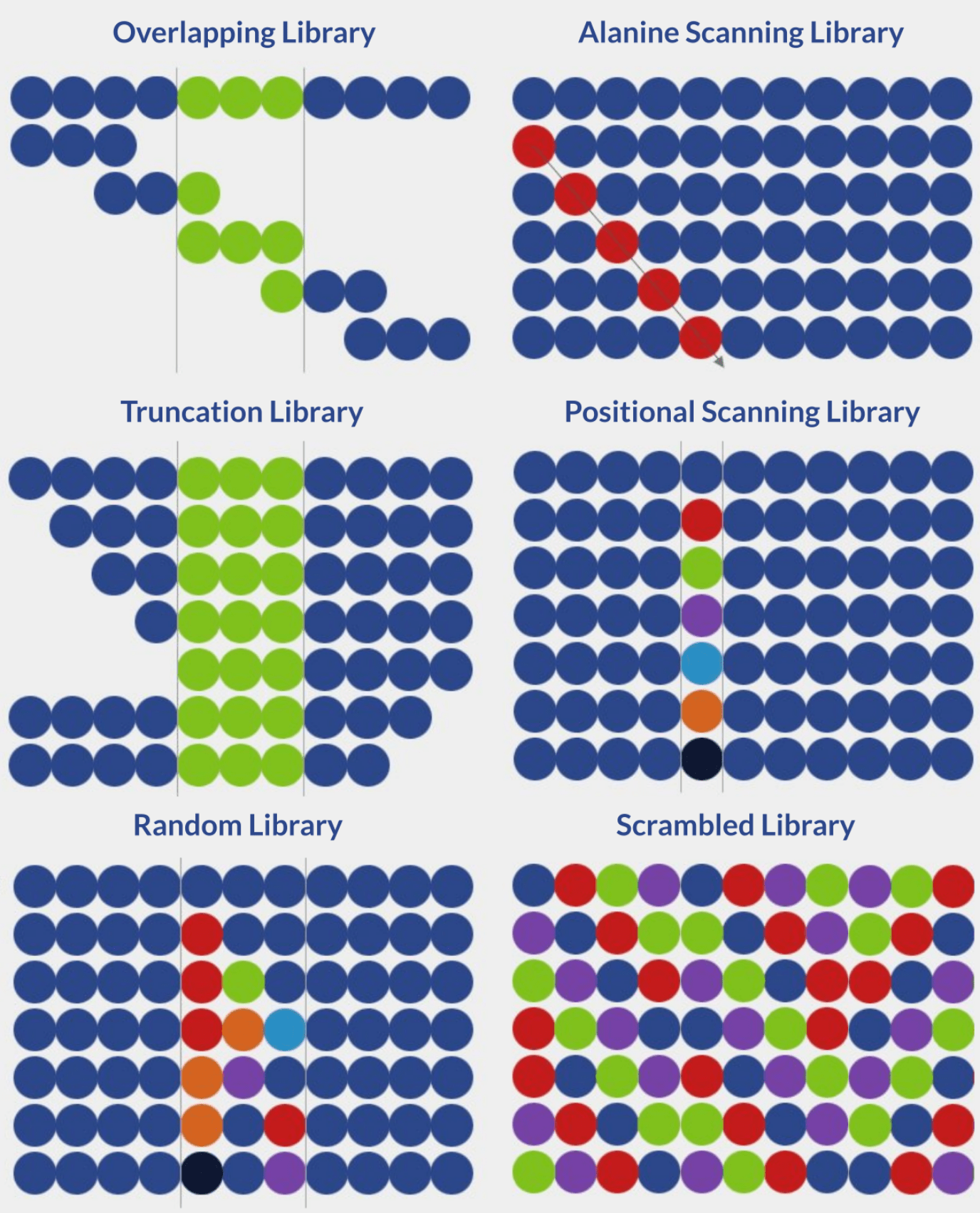
Overlapping peptide libraries
Overlapping libraries are ideal for screening linear or continuous epitopes. The library design process is defined by two parameters:
- Peptide length: the optimal length of an epitope is between 8 and 20 amino acid residues because epitope-paratope interactions typically require between 6 to 12 residues.
- Offset number: it dictates the degree of overlap between sequences. Ideally, it should be 1/3 of the peptide length.
The careful selection of these parameters will determine how informative the experiment will be. Although small peptides reduce the cost of library synthesis, they are also less likely to interact efficiently with paratopes.
Low offset values and short peptide lengths result in libraries with numerous sequences. In contrast, high offset values and long peptide lengths lead to small and low diversity libraries. The choice of the ideal parameters should be determined by pre-existing information regarding the bioactivity of the native protein used as the framework for peptide library design.
The most common applications of overlapping peptide libraries include:
- Epitope mapping (i.e., scanning of antigen sequences for epitopes)
- Paratope mapping (i.e., scanning the variable regions of antibodies for paratopes)
- Determine potential substrates of enzymes
- Identify T-cell epitopes
- Identify B-cell epitopes
- Identify which residues within an epitope are vital for activity
Alanine scanning peptide libraries
In alanine scanning libraries, all amino acid residues in a specific peptide sequence are sequentially substituted by alanine residues, one at a time. These sequential substitutions result in changes in bioactivity, allowing the accurate determination of the importance and role of each residue in epitope-paratope interactions. Alanine is typically used in this type of screening because it is chemically inert and the smallest natural amino acid.
The most common applications of alanine scanning peptide libraries include:
- Discovery and study of functional epitopes
- Elucidation of protein binding sites
- Evaluation of an enzyme’s potential substrates
- Identify key residues for protein or cell engineering
Truncation peptide libraries
Truncation peptide libraries are built by systematically removing terminal amino acids from a peptide sequence from the N-terminus, C-terminus, or both. Systematic truncation leads to systematically shorter peptides. In turn, the screening of these peptides allows for gauging the influence of regions surrounding key residues on peptide bioactivity. These libraries are typically produced and screened after key residues have been identified via alanine scanning.
The most common applications of truncation peptide libraries include:
- Identification of the minimal epitope length required for peptide bioactivity
- Elucidation of the minimal binding site
- Identification of sequences with enhanced proteolytic stability
- Identify sequences that play a major role in aggregation (e.g., in antibody drugs)
- Evaluation of the extent of metabolic degradation of peptide drugs
Positional scanning peptide libraries
When designing positional scanning libraries, each amino acid residue is systematically replaced by all other amino acids, one at a time. These libraries are important for peptide sequence optimization because they allow the identification of residues that enhance or undermine peptide bioactivity.
The most common applications of positional scanning libraries include:
- Optimization of peptide binding sites
- Improvement of T-cell epitopes
- Enhancement of enzyme activity against a substrate
Random peptide libraries
The design of random peptide libraries is simple. Using a shotgun approach, selected residues are randomly and simultaneously substituted with each of the 20 natural amino acids. The random nature of amino acid substitution allows the generation of vast peptide libraries with many variations. In turn, the screening of these sequences allows the identification of novel bioactive peptides.
The most common applications of random peptide libraries include:
- Generation of highly active novel peptide sequences
- Optimization of existing peptides with known bioactivity profiles
- Identification of biomarkers for drug discovery
Scrambled peptide libraries
Among all the major strategies for the design of peptide libraries, scrambled libraries provide the highest degree of variation. The library is generated through sequence permutation, where the amino acid residues are mixed to form all possible amino acid combinations. Due to its vast diversity, scrambled libraries are ideal for peptide sequence optimization.
The most common applications of scrambled peptide libraries include:
- Identification of bioactive peptide mimotopes (peptides that mimic the original epitope by binding to the same antibody)
- Negative controls to prove sequence specificity
- Identification of key residues and peptide sequence optimization
References
- Bozovičar, K. and Bratkovič, T. Evolving a Peptide: Library Platforms and Diversification Strategies. Int J Mol Sci. 2020; 21(1): 215. doi: 10.3390/ijms21010215
- Gray, B. P. and Brown, K. C. Combinatorial Peptide Libraries: Mining for Cell-Binding Peptides. Chem Rev. 2014; 114(2): 1020–1081. doi: 10.1021/cr400166n
- Quartararo, A. J., et al. Ultra-large chemical libraries for the discovery of high-affinity peptide binders. Nat Commun. 2020; 11: 3183. doi: 10.1038/s41467-020-16920-3